Methods
Collection of DNA barcodes
The DNA extraction method which we have adopted for this project is mostly non-destructive in nature.
This non-destructive method enables DNA extraction which does not involve homogenizing or removing any body part from the specimen.
After analyses of DNA sequences, the morphology of same specimen can be fully examined.
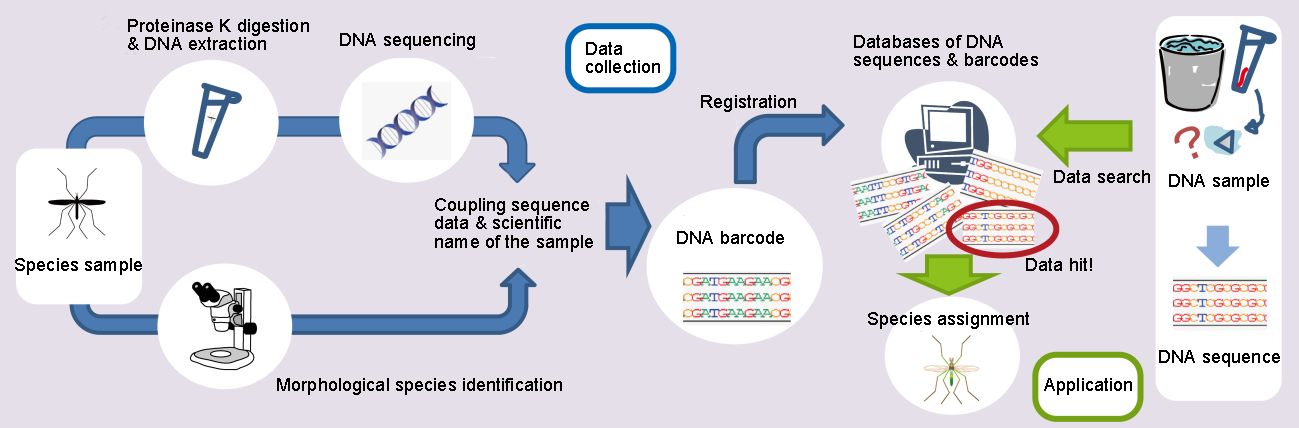
A DNA barcode includes information of a DNA sequence and a scientific name of species. The DNA sequence is determined by extracting DNA from a species sample using proteinase K and the scientific name of the sample is identified by professional morphological examination. DNA barcodes are registered on databases of DNA sequences and barcodes which are used to identify species of unidentified samples by matching DNA sequences.
Advantage of non-destructive extraction in morphological examination
- Simple and quick method of preparation for morphological examination
In the conventional method of preparation for light microscopy, specimens are usually cleared in 10% KOH solution in order to achieve sufficient transparency (Sasa & Kikuchi 1995). In such cases, we must separate the wings from the body, as they will undergo serious damage and deformation by application of KOH treatment.
Non-destructive DNA extraction also results in good transparency, while damaging the wings to a minimal degree. As such, it removes the necessity to dissect the wings, with the consequence that when this method is applied, preparation for morphological examination is both simpler and quicker when compared with the conventional method. However, the optimal extraction time suitable for medium-sized specimens is too short for large specimens while it is too long for small specimens. Therefore, adjusting the extraction time to the sample size becomes an issue, much as is the case with KOH treatment.
- Possible DNA resource for re-extraction
After DNA extraction, we can directly transfer the specimens into glycerol dripped onto a depression slide for microscopic examination. Glycerol works as a substitute for the Hoyer’s solution or Canada balsam which are employed in the conventional observation method. This means that watery reagents which result in the loss of DNA are not needed using our method. As the specimens are transferred from glycerol to 99.5% ethyl alcohol after observation, the remaining DNA in the specimens will be ready for re-extraction.
From extraction of DNA to sequencing DNA segments
- Sample preservation
Put the whole specimen into a 1.5ml microcentrifuge tube filled with 99.5% ethanol.
- Extraction of DNA
-Remove ethanol and dry the specimen
-Add proteinase K and lysis buffer to the specimen (do not homogenize)
-Incubate at 56 °C for 3 hours or more (overnight)
-Retrieve the specimen and preserve in 99.5% ethanol
-Supernatant is used as the template for the following step of PCR
- PCR (Polymerase Chain Reaction)
-Partial sequence of the cytochrome c oxidase subunit I (COI or CoxI) is amplified by PCR with DNA polymerase
-Amplicon size is about 700 bp (658 bp without primer region)
-COI is the standard gene for DNA barcoding of animals
-Primer sets used for this project (Folmer et al. 1994):
LCO1490 (5'-GGTCAACAAATCATAAAGATATTGG-3')
HCO2198 (5'-TAAACTTCAGGGTGACCAAAAAATCA-3')
- Sequencing
-The purified PCR product is directly subjected to Sanger sequencing
-Forward and reverse sequences of the gene are analyzed for every specimen to obtain a consensus sequence
- Analysis of sequences
-Trim primer sequences and acquire a consensus sequence of 658 bp for each specimen
-After morphological species identification and molecular phylogenetic analysis, the sequence is recorded as a DNA barcode for the species.


